You are here
May 23, 2023
Human pangenome boosts accuracy and reflects diversity
At a Glance
- Researchers compiled a set of reference human genome sequences—a “pangenome”—that represents the breadth of human genetic diversity better than a single genome.
- The new pangenome could aid in discovering new genetic variations and ensure that the results of genetic research apply to a broader range of people.

Genetic differences between people can cause or alter the severity of various diseases and influence the effectiveness of treatments. Scientists identify such genetic variants by comparing an individual’s genome sequence to a standard, which is known as a reference genome.
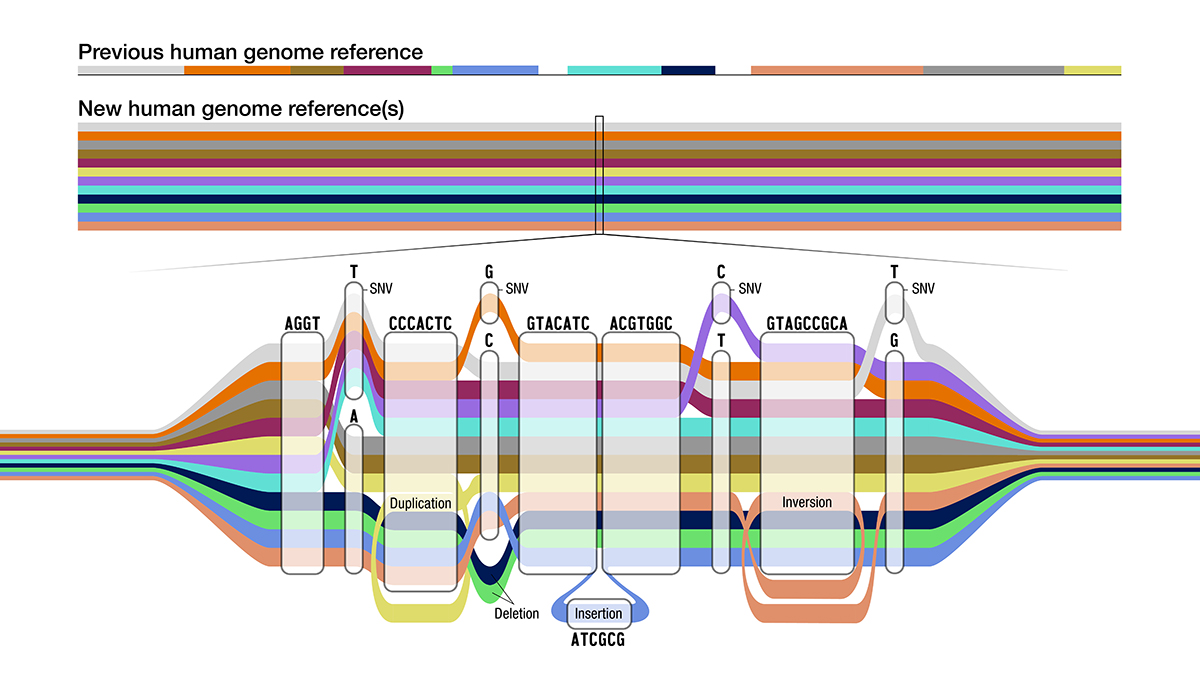
A reference genome is created by assembling parts of the genomes of many different people into a single sequence. The original reference genome was developed by the Human Genome Project two decades ago. It has been continually updated as genome sequencing has become more accurate and more data became available. But a single reference genome can’t represent the genetic diversity of the human species. In particular, larger genetic variations, known as structural variations, are difficult to identify using a single reference genome.
An NIH-funded consortium has developed a reference “pangenome” that represents more human genetic diversity. To do so, they assembled genome sequences from 47 people. About half had African ancestry. Most of the rest were from Latin America and South and East Asia. Only one had European ancestry. Using advanced computer algorithms, the researchers aligned the corresponding sequences within the various genomes.
The pangenome resembles a transit map, with different lines representing each component genome. The lines overlap where the sequences match and branch out where the sequences diverge. A first draft of the pangenome was published in Nature on May 10, 2023. Four companion papers were published as well.
To estimate the completeness of the genomes, the researchers compared them with the first complete human genome sequence released in 2022. On average, the genomes covered more than 99% of the expected sequence. More than 99% of each genome was accurately assembled.
The pangenome captured nearly all human genome variants that have been identified using the existing reference genome, called GRCh38. But it also went beyond the existing reference in several ways. The researchers found more than 1,100 cases of gene duplication in the pangenome that were missing from GRCh38. The pangenome also contains more than 100 million more base pairs—the “letters” of DNA—than GRCh38. Using the pangenome to identify small variants in sequencing data reduced errors by 34% compared with using GRCh38.
Structural variations can be especially hard to detect using a single reference genome. These involve the deletion, duplication, or rearrangement of long DNA stretches. Most of the new base pairs found in the pangenome were in regions that were previously unresolved due to structural variation. The researchers identified previously unknown structural variations at several locations where many such variations are possible. In all, the average number of structural variations identified more than doubled.
The authors note that the published pangenome is only a first draft. The consortium ultimately hopes to produce a more detailed pangenome that incorporates genomes from 350 people. Having a diverse reference may help ensure that future genomic research can benefit people of all backgrounds.
“Since 2000, we’ve had a series of increasingly more accurate representations of one genome,” says consortium member Dr. David Haussler of the University of California, Santa Cruz and the Howard Hughes Medical Institute. “But no matter how accurately you represent one genome, that’s not going to represent all of humanity. Now is a turning point: no longer genomics of the one standard human genome, but genomics for everybody.”
Related Links
- First Complete Sequence of a Human Genome
- Repeats in the Genetic Code Underlie Some Human Variation
- Largest Catalog of Human Genetic Diversity
- Diversity Enhances Genomic Analyses
- Researchers Use New Technique to Decode Goat Genome
- Genetic Misdiagnoses of Heart Condition In Black Americans
- Charting Genetic Variation Across the Globe
- Human Epigenome Map Yields Insights into Development, Disease
- Human Pangenome Reference Consortium
References: A draft human pangenome reference. Liao WW, Asri M, Ebler J, Doerr D, Haukness M, Hickey G, Lu S, Lucas JK, Monlong J, Abel HJ, Buonaiuto S, Chang XH, Cheng H, Chu J, Colonna V, Eizenga JM, Feng X, Fischer C, Fulton RS, Garg S, Groza C, Guarracino A, Harvey WT, Heumos S, … See abstract for full author list … Garrison E, Marschall T, Hall IM, Paten B. Nature. 2023 May;617(7960):312-324. doi: 10.1038/s41586-023-05896-x. Epub 2023 May 10. PMID: 37165242.
Funding: : NIH’s National Human Genome Research Institute (NHGRI), Office of the Director (OD), National Institute of General Medical Sciences (NIGMS), National Library of Medicine (NLM); USDA National Institute of Food and Agriculture; National Science Foundation; Natural Sciences and Engineering Research Council of Canada; Canada Research Chairs Program; Fonds de recherche du Québec; World Premier International Research Center Initiative; Carlsberg Foundation; National Institute of Standards and Technology; Howard Hughes Medical Institute; Oxford Nanopore Technologies; Wellcome Trust; Agencia Estatal de Investigación; NextGenerationEU; Novo Nordisk Foundation; Central Innovation Programme for SMEs; German Network for Bioinformatics Infrastructure; German Federal Ministry of Education and Research; European Commission, Innovative training network; Ministry of Education of Taiwan.